UreA, a urea transporter belonging to unique class of fungi and plant urea transporters.
We have carried out the functional characterization of the UreA permease, and the study of its regulation at the transcriptional level. The analysis of the transporter structure-function relationship, by combining strategies of mutagenesis and structural homology modeling, allowed us to identify key residues involved in its functionality and the binding and translocation of the substrate. Besides the contribution of these studies to the knowledge of the biology of fungi and plants in general, and of this family of transporters in particular, we expect them to be a starting point for the development of strategies for a more efficient use of urea as a fertilizer.
Characterization of cis elements and trans factors that regulate the intracellular traffic of UreA
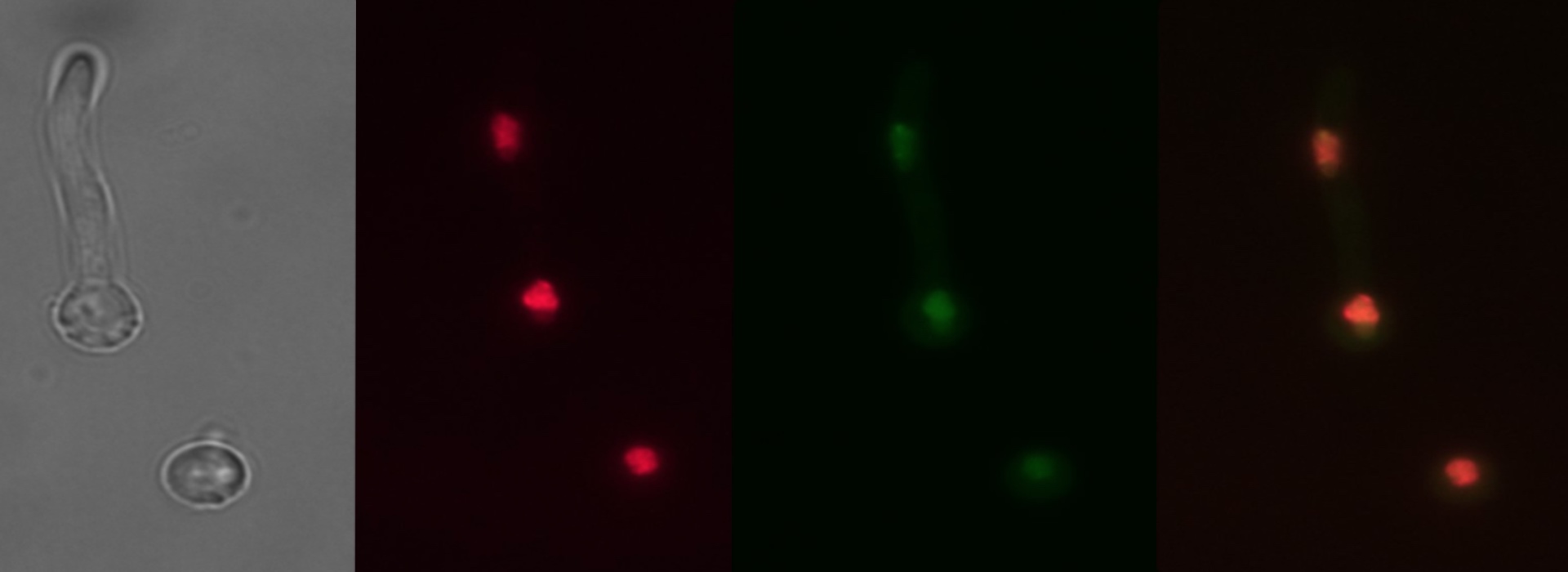
A main aspect in the maintenance of cell homeostasis and stress response is the regulation of the repertoire of transporters present in the plasma membrane (PM), which are responsible for the absorption and extrusion of nutrients, as well as of other small molecules. We aim to contribute to the understanding of the regulation of the intracellular traffic of transporters, by using UreA as a model. We have identified a number of cis-acting elements in the C-terminal domain of UreA, as well as some trans-acting factors (e.g. the HulA ubiquitin-ligase and the ArtA adaptor) involved in the endocytic-mediated degradation process of UreA. We have also identified a putative receptor for cargo proteins (AnErv14) in the endoplasmic reticulum, which would take part in regulating UreA´s sorting to the PM.
A pair of non-optimal codons is necessary for the correct biogenesis of UreA
ureA and its orthologs in the other Aspergillus species possess two conserved, non-optimal codons that code for amino acids located exactly on the border between the unstructured N-terminus and the first transmembrane segment. Mutating these codons by their optimal synonyms leads to a significant decrease in UreA levels, which is not due to a decrease in gene transcription levels or an increase in protein degradation. We then attribute the low protein levels to a defect in its synthesis which, given the early location of the codons, should affect the early stages of the process. Our hypothesis is that the synonymous mutant lacks a translational pause, encoded in the mRNA, necessary for the correct interaction with SRP, the signal recognition particle, that distinguishes nascent polypeptides destined for the endoplasmic reticulum membrane. We are currently studying the UreA-SRP interaction to verify this hypothesis.

Chromatin structure and function in Aspergillus nidulans
Despite the “universality” of the general characteristics of chromatin, there are differences in its structure and function even within organisms of the same genus. In the genus Aspergillus, we have detected important differences at the level of the set of the “canonical” histones and their variants. These differences, beyond the interest they arise from the point of view of the Aspergilli basic cell biology, could lead to the identification of therapeutic targets to fight against fungal infections, the incidence of which has recently increased dramatically, especially among immunocompromised patients. In this context, we study the role of H4E, a variant of histone H4 present exclusively in ascomycete fungi. This would be the first histone H4 variant described so far.
Aspergillus nidulans´ EMC: characterization and identification of clients
Integral membrane proteins (IMPs) are transmembrane proteins that participate in a wide variety of biological processes, such as solute transport and signal transduction. These proteins are encoded by ~25% of protein-coding genes in all organisms and defects in their function are associated with various diseases and drug sensitivity, highlighting their importance. An abundant and widely conserved complex has been identified in eukaryotes, called the ER membrane complex (EMC), which has been implicated in the biogenesis of these proteins. Loss of EMC function has been associated with defects in cholesterol homeostasis, autophagosome formation, and neurological defects, among others. Therefore, understanding how this complex works, as well as identifying its client proteins (and therefore the selectivity of the complex), is of great importance. This is why we seek to contribute to its knowledge, through its genetic and biochemical characterization in the model ascomycete fungus Aspergillus nidulans.


